The genome is identical in every cell of the body. However, this tightly-packed genetic material isn’t always folded into the same shape in each cell. The folding pattern can lead to variations in which genes are activated to make proteins.
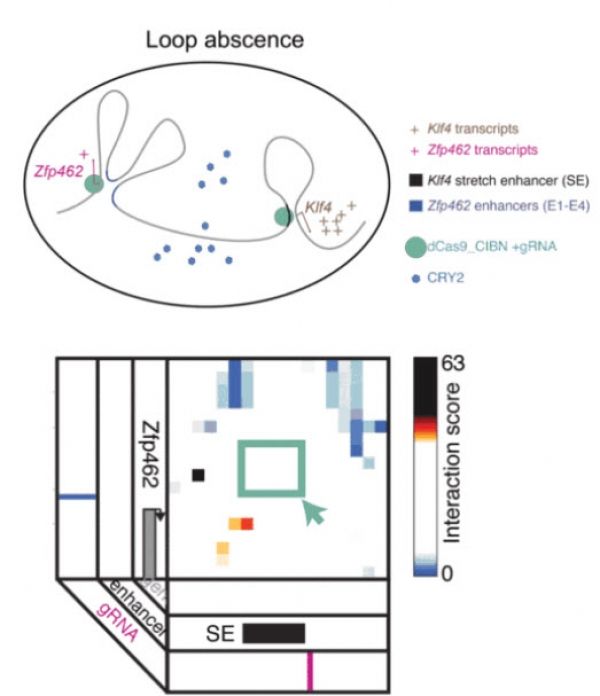
A genome can be thought of as a beaded string, with each bead representing a gene. Reporting in Nature Methods, Jennifer Phillips-Cremins, an assistant professor in Penn Engineering’s Department of Bioengineering, led a team in using light to force both ends of that string together, folding it into specific shapes so that certain genes are in direct physical contact with each other. By controlling which genes are touching, Phillips-Cremins and colleagues hope to determine how different configurations lead to different combinations of genes that are expressed in the body.
This field of genomic shape manipulation is known as “3D Epigenetics,” and Phillips-Cremins is one of the researchers at its forefront. Her team’s light-triggered folding method, known as light-activated dynamic looping (LADL), can fold genomes into specific loops in a matter of hours. The loops are temporary and can be easily undone. Since prior research from the Phillips-Cremins lab indicates that these looping mechanisms may play a role in some neurodevelopmental diseases, this speedy new folding tool may one day be of use in further research or even treatments.
Read more at University of Pennsylvania
Image: LADL combines CRISPR/Cas9 and optogenetics to bring two distant points in a linear gene sequence into physical contact, forming a folding pattern known as a “loop.” Looping interactions influence gene expression, so the researchers envision LADL as being a powerful tool for studying these dynamics. CREDIT: University of Pennsylvania