The search for effective biological tools is a marathon, not a sprint, even when the distances are on the microscale. A discovery at Rice University on how engineered communities of cells communicate is a long step in the right direction.
The Rice lab of synthetic biologist Matthew Bennett has designed a set of transcriptional circuits that, when added to (and expressed by) the genomes of single-cell microbes, allows them to quickly form a network of local interactions to spur collective action, even in large communities.
Research published in Nature Chemical Biology shows engineered strains of Escherichia coli transmitting signals down a bacteria-filled corridor and coordinating their actions. The ability to do so could lead to engineered microbes that treat conditions in gut microbiomes or communicate with bioelectronics.
“Cells often use chemical signals to communicate and relay information to each other,” Bennett said. “However, chemical signals have a limited range. After they leave the cell, they diffuse through whatever medium the cells are in, and that can only go so far.
“In this study we looked at a previous system we built that uses two different strains and different types of communication between them to study how, once we increase the size of the colony containing these strains, it would react,” he said.
Read more at: Rice University
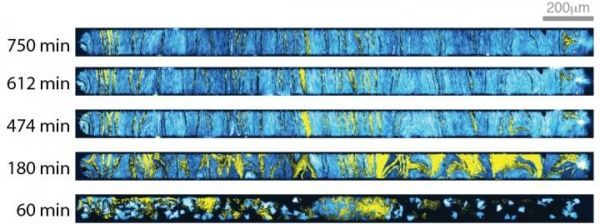
Representative fluorescence images of cells growing in the open microfluidic device developed at Rice University show how transcriptional circuits allow single-cell microbes to form networks that spur collective action, even in large communities. The spatial arrangement of two strains of color-coded cells eventually stabilizes. (Photo Credit: Courtesy of The Bennett Lab/Nature Chemical Biology)