New structure shows how virus envelope protein hijacks cell-junction protein and promotes viral spread. Findings could speed the design of drugs to block severe effects of COVID-19.
Scientists at the U.S. Department of Energy’s (DOE) Brookhaven National Laboratory have published the first detailed atomic-level model of the SARS-CoV-2 “envelope” protein bound to a human protein essential for maintaining the lining of the lungs. The model showing how the two proteins interact, just published in the journal Nature Communications, helps explain how the virus could cause extensive lung damage and escape the lungs to infect other organs in especially vulnerable COVID-19 patients. The findings may speed the search for drugs to block the most severe effects of the disease.
“By obtaining atomic-level details of the protein interactions we can explain why the damage occurs, and search for inhibitors that can specifically block these interactions,” said study lead author Qun Liu, a structural biologist at Brookhaven Lab. “If we can find inhibitors, then the virus won’t cause nearly as much damage. That may give people with compromised health a much better chance for their immune systems to fight the virus successfully.”
Scientists discovered the details and developed the molecular model using one of the new cryo-electron microscopes at Brookhaven Lab’s Laboratory for BioMolecular Structure (LBMS), a new research facility built with funding from New York State adjacent to Brookhaven’s National Synchrotron Light Source II (NSLS-II).
Read more at DOE/Brookhaven National Laboratory
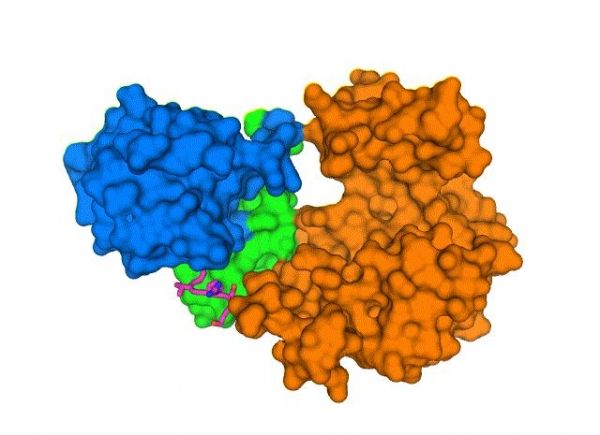
Image: New structure shows how the COVID-19 virus envelope protein (E, magenta sticks) interacts with a human cell-junction protein (PALS1, surfaces colored in blue, green, and orange). Understanding this complex structure, which was solved using a cryo-electron microscope at Brookhaven National Laboratory, could lead to the discovery of drugs that block the interaction and, potentially, the most severe effects of COVID-19. (Credit: Brookhaven National Laboratory)